RADWASTE MANAGEMENT
Toxic avengers
18 March 2006Life adapts to survive in the most forbidding conditions – even in former uranium mining and production sites. If man could understand how microbes handle radionuclides, he might one day harness a natural force for remediation. By Johannes Raff and Sonja Selenska-Pobell
Many microbes are able to tolerate radionuclides and other hazardous elements in their environment. Adaptation and detoxification mechanisms allow them to resist very high concentrations of toxic elements without being permanently affected. These mechanisms represent an excellent prospect for the development of innovative remediation strategies.
Decades of uranium mining and production in eastern Europe, including Saxony and Thuringia in Germany, has left its mark in these regions, including uranium polluted soil, subsoil and seepage water at former production and dumping sites. Because of uranium’s radioactivity and toxicity, adequate measures have to be taken to exclude its potential effects on mankind and the environment. Processes like weathering, leaching, and anthropogenic activities led to a mobilisation of uranium and therefore a higher risk of accumulation of uranium in the food chain. Besides the general challenge to remediate these sites, questions of uranium contamination have also become of concern for the production of drinking water in Germany. In both cases the complete and selective removal of heavy metals is desirable. Conventional methods for cleaning contaminated water such as precipitation, coagulation and membrane-based processes are expensive and less effective at low metal concentrations. Many studies on immobilisation of dissolved uranium have also been conducted with inorganic and organic sorbents and with different kinds of biocomponents, such as lichen biomass, bacteria, algae, fungi, plants and animal biopolymers. These materials can be very expensive and often bind a wide range of metals, decreasing their efficiency for a particular metal such as uranium, and mostly cannot be recycled.
A new prospective strategy emerged from the cooperation of researchers from the Institute of Radiochemisty, Forschungszentrum Rossendorf, the Institute of Materials Science, Technical University Dresden and the group Functional Coatings, Association for the Advancement of Biological, Medical, and Environmental Technologies, Dresden (GMBU, Gesellschaft zur Förderung von Medizin-, Bio- und Umwelttechnologien eV). The idea was to construct a selective and reusable biocomposite material and to this end, metal binding bacteria, recovered from the uranium contaminated sites, were embedded in a SiO2 matrix by sol-gel techniques.
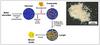
What makes bacterial biomass more prospective than other biomass? Bacteria are, after archaea, the oldest group of organisms on our planet, are the most common organisms on Earth and are more successful in colonising extreme and hostile environments than every other kind of life. The reason for their success is the ability to adapt to a large variety of environmental conditions, thus bacteria are able to interact in many different ways with radionuclides such as uranium. The biosorption, bio-accumulation, biotransformation (reduction and oxidation), biomineralisation and microbially-enhanced chemisorption mechanisms are well studied (see Figure 1).
Biosorption means the chemical sorption of radionuclides by complex formation with cellular ligands on the cell surface. In contrast, bioaccumulation refers to the processes by which the metals are accumulated inside the cell. The reduction or oxidation of radionuclides are summarised as biotransformation, while biomineralisation describes the formation of insoluble precipitates. The last mechanism of possible microbe-radionuclide interaction is the microbially-enhanced chemisorption, covering the inclusion of metal cations in inorganic precipitates. All these mechanisms strongly influence the mobility of the metals in the environment.
The preferred mechanisms for removing dissolved metals is biosorption because of the potential for reusing a possible filter material and a high binding capacity combined with a high binding selectivity. For the discovery, therefore, of suitable bacteria, the most promising method is to investigate bacteria recovered from sites that are highly contaminated with the radionuclide one wants to remediate.
In recent years, many microbial (bacterial and archaeal) communities of former uranium mining and production sites were studied at the Institute of Radiochemistry. As a first step, bacterial communities were characterised by direct molecular methods. These methods are based on the isolation of DNA from soil or water samples and direct analysis of the microbial 16S rRNA genes. These genes encode the 16S rRNA molecules, which are structural parts of the small ribosomes subunit, the cellular components of protein synthesis. The method allows the identification of novel not yet cultured bacteria that can even be predominant in the environment. This is a big advantage in comparison to the enrichment culturing methods, which are based on our limited knowledge about the life necessities of only a small part (about 1%) of the existing organisms on Earth.
Our results demonstrated very high and site-specific diversity in the different samples. Interestingly, several bacterial groups, including also novel lineages, seem to be characteristic and predominant in the heavy metal contaminated water and soil samples. Therefore one can speculate that there are pollutant-resistant bacteria that protect the non-resistant bacteria in the investigated consortia. Besides this, synergistic effects inside the consortia may play an important role for the survival of the whole community.
Applying the enrichment culture methods in parallel, particular representatives of the analysed bacterial communities were recovered and their resistance to and interaction with radionuclides was investigated to estimate their potential to function as an effective binding matrix. The understanding of the microscopic and macroscopic processes of the radionuclide/microbe interaction is of fundamental interest. Different spectroscopic, microscopic and biochemical methods were used to investigate where, how and in which amounts radionuclides interact with the microbes. From this, several interesting isolates were found including bacteria that are able to enzymatically reduce soluble uranium(vi) to the insoluble uranium(iv). These bacteria, belonging to the sulfate-reducing group are slow-growing and often used to prevent migration of uranium in contaminated soil by immobilisation, or to remediate contaminated water by uranium precipitation.
As the bacterial activity in soil is not necessarily always uniform and the conditions may change, reoxidation and therefore mobilisation of the uranium may occur. Furthermore, for the remediation of contaminated water, nutrients must be fed to the bacteria, and separation of the uranium precipitate from the biomass is difficult. Thus after removal of the uranium, it has to be mobilised again for a reuse of the biomass or the uranium precipitate has to be disposed together with the biomass.
Other fast-growing isolates belonging to the actinobacteria group are able to accumulate large amounts of uranium. But as the accumulation inside cells retards the release of uranium, which is necessary for the reusability of the material, these bacteria are less interesting for bioremediation techniques. Several bacteria belonging to the genus bacillus are more promising for remediation application. These bind uranium selectively and reversibly in large amounts on the cell surface and on the surface of spores. Bacilli are widespread gram-positive bacteria, which are able to produce endospores under certain conditions, allowing the cells to outlast harsh environmental conditions (such as lack of nutrients, water, or heat) and are therefore cell forms important for bacterial survival.
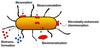
A researcher from the Institute of Radiochemistry found that intact cells, spores and a proteinaceous surface layer (S-layer) of several bacilli selectively bind large amounts of uranium by biosorption. The S-layers are prevalent surface structures of bacteria and archaea which form highly regular protein lattices, covering the whole cell. Therefore the S-layer serves as an interface between the bacterial cell and its environment. These protein envelopes show different kinds of lattice symmetries (oblique, square or hexagonal), and are 4-30nm thick with regularly distributed pores (overall porosity 30-70%) of a size ranging from 2-8nm (see Figure 2).
“” |
By binding the uranium on its surface, the cell may prevent itself from getting affected by the toxicity of the uranium |
S-layers account for 10-25% of the total protein of a cell, are able to self-assemble in monolayers on many different surfaces and possess a high content of charged amino acids. Although S-layers have been known for more than 50 years, apart from the function of S-layers on pathogenic bacteria as virulence factor, their role in nature remains unclear in most cases. Several experiments have proved that they may work as carrier of exoenzymes, as a protective coat against parasitic bacteria, as a structure involved in cell adhesion and surface recognition, as a barrier for toxic elements or as a template for biomineralisation.
The first intensive studies were carried out using the bacillus sphaericus isolate JG-A12. This strain was recovered from the Haberland uranium mining waste pile situated near the town of Johanngeorgenstadt, Saxony, Germany. The cells, spores and S-layer protein are able to bind 64.0, 109.5 and 19.5mg uranium per g dry weight (dw). The error range of these measurements was 4-9%. Interestingly the surface layer of the waste pile isolate B sphaericus JG-A12 possesses in comparison with the S-layer of a reference strain B sphaericus NCTC 9602 and with a reference protein (bovine serum albumine) a significantly higher affinity to uranium in solution. This finding is very astonishing because of the similar molecular structure of the S-layer proteins of the B sphaericus strains JG-A12 and NCTC 9602. Whereas the S-layer of B sphaericus JG-A12 is able to bind almost all uranium in environmental relevant concentrations of 5mg/l and below at pH 4.5, the other two proteins showed decreasing binding capacities with decreasing uranium concentrations.
The investigation of posttranslational modifications of both S-layer proteins revealed a six-fold higher phosphorus content of the S-layer from the waste pile isolate in comparison to the S-layer of the reference strain, whereas the amino acid composition is nearly the same. These results may reflect an adaptation of B sphaericus JG-A12 to its uranium contaminated environment. By binding the uranium on the surface, the cell may prevent itself from getting affected from the toxicity of uranium.
BINDING THE BIOMASS
Since pure biomass is not suitable for a technical application such as the removal of contaminants from water, the biomass has to be immobilised reliably. This was accomplished by the complete encapsulation of cells, spores and S-layers in a biocomposite material (biocer) by sol-gel techniques (see Figure 3).
Silicon- or metal oxide sols were then produced by hydrolysis of their corresponding alkoxides. After the formation of a stable nano-crystalline dispersion, the alcohol was removed and continuously substituted with water. After mixing the sol with the biocomponent, changes of the pH value or increases in temperature lead to a fast gelation process. Subsequently performed drying steps generated porous biocers well suited for use as filter materials. Using sol-gel techniques bulk particles of different sizes, and thin coatings on, for example, glass carriers can be produced. The immobilisation of biomass in a porous but mechanically robust material, has two advantages: the high accessibility of the ligands on the surface of the biomass and the reusability of the filter material in bioreactors. During immobilisation the biocomponents retrain their conformation and metal binding capacity. Although the sol-gel protocols allow keeping biomass alive and enzymes active during the immobilisation process, in the case of this application the binding process needs no metabolically active bacteria or spores. For the production of biocomposite materials, either cells, spores or S-layer protein of B sphaericus JG-A12 were embedded in the porous silicate matrix (see Figure 4).
The results of uranium sorption tests of free and immobilised bio-components of JG-A12 under acidic conditions (pH 4.5), showed that the binding capacity of the cells and S-layer was not negatively influenced by the immobilisation procedure. Additionally, the biocers can be regenerated for multiple application by washing the material with complexing reagents such as citric acid, which removes bound uranium completely. But the overall binding capacity of the biocers is 1.4-8.6-fold lower than the same dry weights of the free biocomponents. Thus cell biocers bind 18.6mg uranium, spore biocers 12.8mg uranium, S-layer biocers 14.2mg uranium and the pure xerogel 13.5mg uranium by g dw. There are two reasons for that: with all biocers the biomass content is very low at 20% because of the danger of decreasing mechanical stability, and in the case of the spore biocer, the accessibility of the bioligands is reduced after immobilisation. To overcome these restrictions, new methods for the immobilisation will be tested and biomass with higher uranium binding capacities with an equally high affinity for uranium will be used.
The screening of several bacillus isolates from the Haberland waste pile showed the presence of S-layers in 9 of 21 isolates. Preliminary uranium binding experiments in acidic conditions showed binding capacities of 11.6±2.5 to 229±25 mg uranium by g dw for intact, S-layer carrying cells and very high binding capacities of 42.5±2.5 to 291.5±4.1 mg uranium by g dw for the purified S-layer proteins.
Former studies on uranium binding by cells, spores and S-layer proteins from the two B sphaericus strains JG-A12 and NCTC 9602 proved the role of carboxyl and phosphate groups for the biosorption of uranium. Interestingly also, for most new S-layer proteins there seems to be a correlation between the amount of phosphate present in the S-layer proteins and the amount of bound uranium.
Developing improved cleanup strategies for contaminated water, cells and spores is useful for the removal of uranium from contaminated seepage or other waste water, whereas for hygenic reasons S-layers are especially suitable for cleaning drinking water. Furthermore, this offers the possibility to increase binding capacity and binding selectivity by chemical and biomolecular protein modifications, for instance by adding specific metal binding peptides or binding relevant amino acids. The latter were identified by sorption experiments with modified and non-modified proteins and by spectroscopic analysis.
Besides the screening for a more selective metal binding S-layer with high binding capacities and the protein modification, the combination of biomass and therefore the use of synergistic effects will be a further promising way to improve binding properties of the biocomposite material, up to the point of a nuclide-selective binding of elements.
Finally, one can assume, that although bacteria are one of the smallest and most simply organised organisms on Earth, they are an unbelievably adaptive, metabolically diverse and effective group of creatures. Many of them are able to resist the most forbidding environmental conditions by adaptation. Understanding the underlying mechanisms of the selective and reversible binding of several metals by surface structures would allow the development of new and innovative bioremediation strategies and their application in nanotechnology.
Author Info:
Johannes Raff and Sonja Selenska-Pobell, Institute of Radiochemistry, Forschungszentrum Rossendorf, PO Box 510119, 01314 Dresden, Germany
| By binding the uranium on its surface, the cell may prevent itself from getting affected by the toxicity of the uranium |